Since 2006, an unknown plague upon western honey bee communities has caused populations to plummet, putting the species in danger of extinction. Beekeepers and agricultural scientists named this mysterious illness Colony Collapse Disorder, as it causes large portions of worker bees to leave the hive and isolate themselves before dying. While there are a whole host of pathogens and illnesses known to be harmful to honeybees--including the modern problem of acute pesticide poisoning--no single pathogen has been identified as a probable cause of the startling phenomenon.
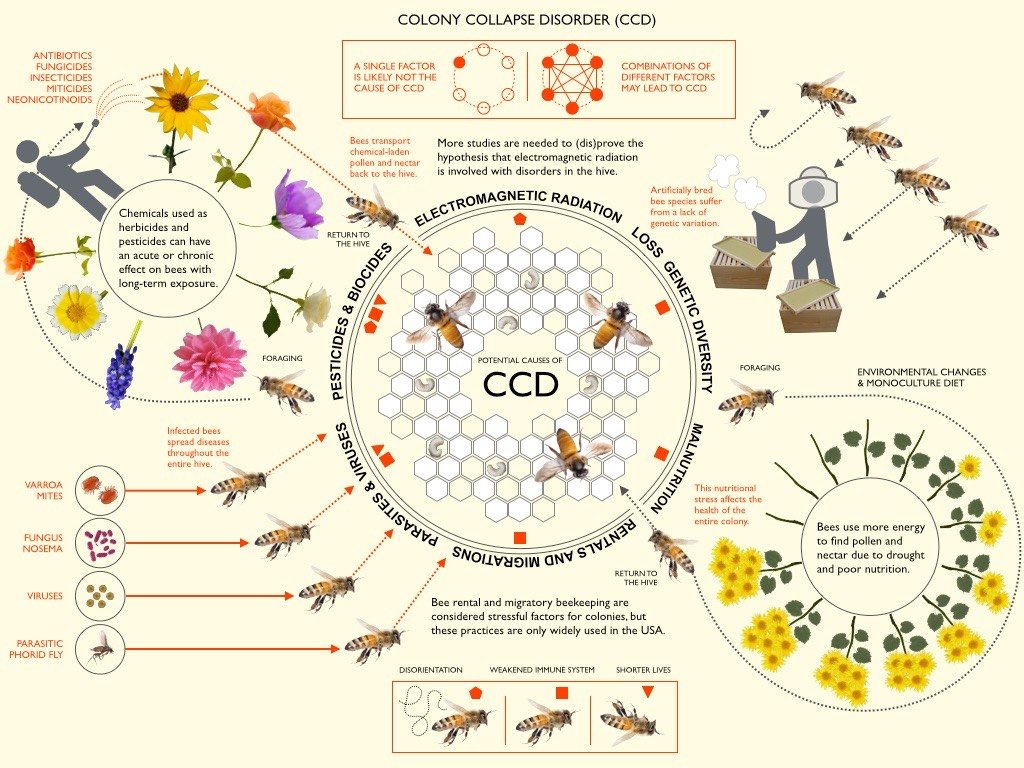
Consequently, researchers from the Agricultural Research Service and University of Illinois conducted a study exploring possible genetic explanations. In the study, scientists collected tissue samples from the guts of honey bees either affected or unaffected by CCD, then performed experimental analysis of gene expression across the whole genome. Here, researchers saw that there were no irregularities in the expression of genes associated with immune function and pesticide response for bees affected by CCD, despite the fact that pathogenic analysis revealed elevated levels of illness and pesticide exposure in these same bees. Furthermore, they found that CCD bees had significantly higher levels of abnormal DNA sequence coding for ribosomal RNA, a key component of functional protein production.
These results surprised experts, but also introduced the very real possibility that Colony Collapse Disorder could be caused by deleterious mutations in the ribosome, which would leave vulnerable honey bees without the proteins necessary to defend against pathogenic attack. With all this data in mind, researchers propose that a virus within the picorna family, or one that directly attacks ribosomes, could be at the root of the most recent threat against the western honey bee species. Hopefully, future studies can explore this hypothesis further and find better methods for protecting against CCD, especially considering how disruptive the disease could be to sustainable and accessible food production.
Generally, the ribosome is understood to be the cellular mechanism by which DNA instructions are translated into active polypeptides. It is comprised of two main components, commonly known as the small and large ribosomal subunits.
As a whole, the structure of the ribosome is often depicted like this:
Here, this simplified graphic shows the building of peptide bonds in action as the mRNA and tRNA collaborate with the ribosome:
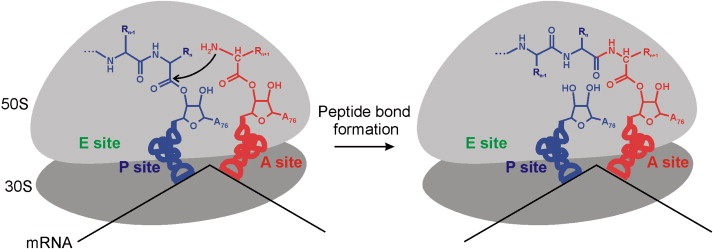 Within the P site, X-ray crystallography analysis has allowed scientists to identify a singular adenine nucleotide responsible for catalyzing bond formation -- which seems impressively simple considering the complexity of other ribosomal activity. Overall, however, biochemists have a highly limited understanding of the ribosome. While basic structural models have been solved, there is still much to learn about the functions and evolution of rRNA molecules. Some studies have found that certain rRNA sequences, particularly the bacterial 16s sequence, have extremely low mutation rates and are highly conserved throughout phylogenies. As a result, they’ve begun collecting databases of rRNA sequence and using these to build phylogenetic trees for bacterial species. Looking forward, additional studies could discover novel methods for examining and understanding the ribosome so that cellular mutations and evolution can be better understood.
Within the P site, X-ray crystallography analysis has allowed scientists to identify a singular adenine nucleotide responsible for catalyzing bond formation -- which seems impressively simple considering the complexity of other ribosomal activity. Overall, however, biochemists have a highly limited understanding of the ribosome. While basic structural models have been solved, there is still much to learn about the functions and evolution of rRNA molecules. Some studies have found that certain rRNA sequences, particularly the bacterial 16s sequence, have extremely low mutation rates and are highly conserved throughout phylogenies. As a result, they’ve begun collecting databases of rRNA sequence and using these to build phylogenetic trees for bacterial species. Looking forward, additional studies could discover novel methods for examining and understanding the ribosome so that cellular mutations and evolution can be better understood.

Johnson RM, Evans JD, Robinson GE, Berenbaum MR. 2009. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera). Proc Natl Acad Sci U S A 106:14790–14795. doi:10.1073/pnas.0906970106.
About the Author
Zoe Brown is a current student of biology and education at Mount Holyoke College. She is originally from Massachusetts, and grew up in a small town by the ocean. In the future, she plans to research and teach biology to the next generation of scientists. For now, however, she lives happily in Western MA with her elderly cat Shiva.
Works Cited
Johnson RM, Evans JD, Robinson GE, Berenbaum MR. 2009. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera). Proc Natl Acad Sci U S A 106:14790–14795. doi:10.1073/pnas.0906970106.
Comments
Post a Comment